GreenLab Course
Principles
GreenLab implementations.
GreenLab main implementations
-
Various GreenLab implementations under different environments are available.
-
They also differ in their specifications, allowing stochastic simulation or not,
fitting or not, retroaction on structure or not.
The following table summarises the implementations and their specifications
| Name | Environment | Stochastic | Retroaction | Diffusion | Language | Main developer | Usage | Url |
| StemGL | Windows/Linux | - | - | Freeware | Matlab/Octave | CIRAD - Amap & BioAgresseur | Education - Research | StemGL |
| GreenScilab | Windows Os | - | - | Freeware | Scilab | Chinese Academy of Sciences - CASIA | Education | GreenScilab |
| Xplo with GreenLab Plug-in | All Os | - | - | Freeware | Java | CIRAD - Amap (Amapstudio) | Research | Xplo |
| dgpSDK libraries | Windows and Linux Os | X | X | Free to partners | C++ | Ecole Centrale Paris | Research | digiplante's software |
| GLOUPS | Windows and Linux Os | X | X | Free to partners | Mathlab | CIRAD- Amap Unit | Research | GLOUPS |
| QingYuan | Windows Os | X | X | Contractual | Qt / C++ | Chinese Academy of Sciences - CASIA | Research | QingYuan |
Some GreenLab implementations and their specifications
StemGL
-
StemGL is a simple tool covering both calibration and simulation aspects of biomass production and allocation. The application limits to continuous growth single stem plants,
involving a limited set of parameters. The tool implements stochastic simulation capabilities. It include physiological advanced processes with density effect, seed size effect,
and biomass reallocation; it offers virtual insights in the plant's growth, including evolution in biomass demand and production, details of the biomass at each date and representation of sink functions.
This tool is running under Matlab and GNU Octave environments. Its iterative form makes it possible to consider coupling this tool with other models, for example physiological ones.
The tool operated with a simple dataset frame of 8 parameter item sets, whatever the species considered. Datasets with adjusted parameters are already available for various temperate
and tropical agronomic plants (beetroot, maize, sunflower, tmato, coffee tree, banana tree …). Climatic variations that limit production can also be introduced.
See StemGL page (http://greenlab.cirad.fr/StemGL/) for download and tutorial.
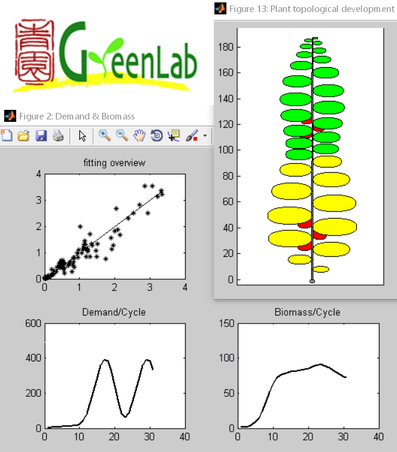
StemGL fitting output example (© CIRAD)
GreenScilab
-
GreenScilab is a toolbox developed by Mengzhen Kang and Qi Riu (CASIA) in the Scilab environment to run the GreenLab model.
GreenScilab can simulate many crop plants, such as tomato, cucumber, chrysanthemum and maize, and these crops have been calibrated with experimental data.
See GreenScilab inline page for download and tutorial.

GreenScilab interface (© LIAMA-CASIA)
Gloups
-
Gloups (GreenLab Universal Plant Simulator) is a toolbox developed by Philippe de Reffye (CIRAD Amap) in the
Matlab environment to run the GreenLab model.
Gloups is a research prototype. It is the most advanced GreenLab implementation, allowing both simulation and fitting.
See Gloups inline page for more information.
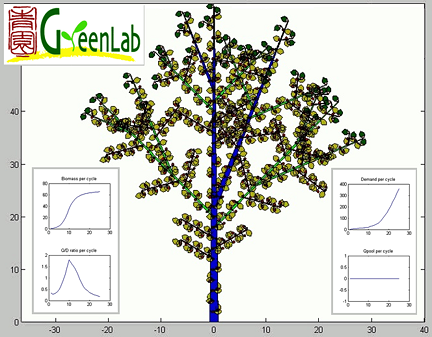
Gloups interface (Snap shot M. Jaeger, CIRAD)
Qinyuan
-
Qingyuan, written by Hua Jing (CASIA) under the Qt programing environment, in C++,
offers an advanced GreenLab implementation with high performance.
It can simulate trees, such as albizia, ginkgo, pine and orange tree....
See http://www.cybernature.com.cn/cPlant/software.html for further information.
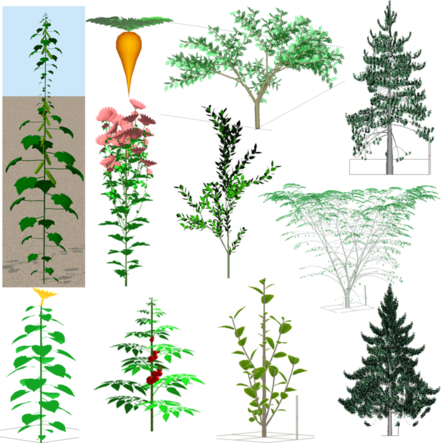
Some QingYuan simulations (H. Jing, LIAMA-CASIA)
PyGMAlion and dgpSDK
-
Developed by Ecole Centrale Paris, in C++, these environments are dedicated to researchers.
PyGMAlion (Plant Growth Modelling Analysis and identification) is a C++ framework embedding modelling, simulation, parameter estimation, and discrete system sensibility analysis tools.
dgpSDK is a library set implementing the GreenLab model with basic applications.
See http://digiplante.mas.ecp.fr/software for further information.
